Scroll to:
Personalized selection of topical antibacterial agents in patients with microbial eczema based on whole genome sequencing data: A prospective сomparative randomized study
https://doi.org/10.25207/1608-6228-2025-32-4-18-32
Abstract
Background. Each individual has a unique skin microbiota that includes a diverse resident and transient community displaying an individualized composition. This study focuses on the identification of skin microbial genomes in patients with chronic microbial eczema.
Objective. To evaluate the clinical efficacy of complex therapy in patients with microbial eczema using personalized selection of topical antibacterial agents while taking into account the determination of antibiotic resistance by whole genome sequencing.
Methods. The prospective сomparative randomized study involved 60 patients with microbial eczema in the exacerbation stage, who were randomly divided into two groups of 30 people each: main and control groups. In the control group (group 2, n = 30) patients were treated according to the Federal Clinical Guidelines. In the main group (group 1, n = 30) patients were also treated with conventional therapy, while topical antibacterial agents were selected in a personalized manner based on antibiotic resistance data. On the 21st day of therapy, an emollient with a probiotic component was added to the treatment regimen for patients of both groups. The patients underwent therapy, observation, and examination at the Department of Dermatovenerology of the Kuban State Medical University of the Ministry of Health of the Russian Federation. The clinical study was performed between December 2023 and December 2024. The observation periods in the study were divided into several stages: before the start of therapy, on days 14, 21, and six months after treatment. In this study, the microbiome of the affected skin areas of the patients was analyzed comparatively, and the effect of topical antibacterial preparation selected on the basis of whole genome sequencing data was evaluated. The laboratory stage of the present study was conducted at two sites: CL Lab Clinical Diagnostic Laboratory (“СL Medicalgroup” LLC) in Krasnodar and Serbalab Genetic Laboratory (“Serbalab” LLC) in St. Petersburg. The Eczema Area and Severity Index (EASI) was used to evaluate the skin pathologic process. Statistical processing of data was performed using the Statistica 12.0 software package (StatSoft, USA) and Microsoft Exel 2010 (Microsoft, USA). Quantitative parameters were statistically described using median and quartiles (Me (Q1;Q3)), along with mean with standard deviation (M ± SD). Differences were considered statistically significant at the error level of p < 0.05.
Results. During exacerbation in patients with chronic microbial eczema, the microbial equilibrium shifted towards an increase in microorganisms Staphylococcus aureus, Clostridioides difficile, Klebsiella pneumoniae, Pseudomonas aeruginosa, Escherichia coli that were present on the affected skin areas, while the number of functional bacteria was limited. A more pronounced tendency to restoration of healthy microbiome in the patients of the main group was observed as soon as the 14th day. By the 6th month of treatment, the patients of the main group experienced a greater decrease in microbial colonization in the foci of microbial eczema lesions than in the control group, whereas the proportion of Bifidobacterium and Lactobacillus functionalis was increased compared to the pre-treatment values by 2.31 and 2.10 times, respectively. In addition, the disease relapse occurred in two patients in the main group (versus five in the control group), which may indicate the higher efficacy of the therapy method proposed in the study.
Conclusion. Whole genome sequencing method identifies the taxonomic diversity of the microbiome. Personalized application of topical antibacterial agent in complex therapy in the main group of patients promotes faster restoration of healthy skin microbiome than in the control group.
For citations:
Lazarev V.V., Tlish M.M., Shavilova M.E. Personalized selection of topical antibacterial agents in patients with microbial eczema based on whole genome sequencing data: A prospective сomparative randomized study. Kuban Scientific Medical Bulletin. 2025;32(4):18-32. https://doi.org/10.25207/1608-6228-2025-32-4-18-32
INTRODUCTION
Chronic microbial eczema (ME) is a common recurrent inflammatory skin disease that significantly worsens the quality of life of patients [1][2]. In genetically predisposed individuals with disorders of innate and adaptive immunity, this disease can be provoked by exogenous and endogenous factors. Dysbiosis and the poor physiologic diversity of the skin microbiota is another factor supporting chronic inflammation on the skin in microbial eczema. The severity of the disease course correlates with the prevalence on the skin of patients of virulent strains of Staphylococcus aureus, contamination of which occurs due to a lack of antimicrobial peptides and dysregulation of innate immunity. In addition, the integrity of the epidermal skin barrier is compromised, skin proteases are activated and superantigens are synthesized, particularly SEA (staphylococcal enterotoxin A), SEB (staphylococcal enterotoxin B), SEC (staphylococcal enterotoxin C) and toxic shock syndrome toxin-1 (TSST 1). The latter activates Th-lymphocytes (Th2) without prior presentation on the surface of antigen-presenting cells and trigger the release of cytokines (IL36 (interleukin), IL17, IL31, TSLP (thymic stromal lymphopoietin; thymic stromal lymphopoietin)), the excess of which leads to systemic toxicity and suppression of the adaptive immune response, which creates a favorable environment for pathogens [3–5].
Microbial eczema is prone to exacerbation, due to an increase in the number of opportunistic and pathogenic microorganisms on the skin. The latter creates prerequisites for the development of autoallergic component that leads to the chronic form of the disease. During exacerbation, acute inflammation of the skin is characterized by itching, dryness, desquamation, as well as the formation of vesicles that quickly turn into wet erosions (serous wells) or serous crusts. In the long course of the disease, patients experience stress, which serves as an additional trigger for the manifestation of inflammatory reactions and activation of opportunistic flora [6][7].
The skin microbiome in chronic microbial eczema undergoes dysbiotic changes marked by an increase in the proportion of microorganisms of various opportunistic families: Staphylococcus aureus, Pseudomonas aeruginosa, Clostridioides difficile, Klebsiella pneumoniae, Escherichia coli, which replace representatives of the functional flora (Staphylococcus epidermidis, Bifidobacterium, Lactobaccilus)1 [8]. The role of nonspecific pathogens such as Proteus vulgaris, Neisseria meningitidis, Neisseria gonorrhoeae, Cl. perfringens, Cl. histolyticum, and Cl. septicum has also been described in the pathogenesis of ME [9][10].
Etiopathogenetic mechanisms of microbial eczema development necessitate the prescription of antibacterial drugs. Courses of systemic and external antibacterial therapy in patients with chronic ME can reduce the contamination of representatives of bacterial pathogenic flora. This may be accompanied by a decrease in inflammatory processes and clinical improvement of eczema [10].
Recently, the increasing resistance of pathogens to antibacterial drugs has become a growing concern. The irrational use of these drugs can lead to the potential development of resistance to systemic and topical antibacterial drugs [11]. The development of antibiotic resistance is conditioned by genetic mechanisms such as acquisition of new genetic information or changes in the expression level of one’s own genes [12][13].
Therefore, the use of molecular genetic diagnostic methods to detect antibiotic resistance is a promising direction. The new molecular genetic method of whole genome sequencing (WGS) facilitates in-depth analysis of microbiome composition. This method serves to identify and genomically characterize archaea, bacteria, fungi, parasites, and viruses without prior identification of a specific pathogen directly from clinical samples. WGS provides an assessment of antibiotic resistance and toxicity genes along with recommendations for probiotic correction for the physician. Unless these data from WGS are taken into account, treatment of microbial eczema may be ineffective with an increase in the number of relapses and clinical deterioration of the disease. Thus, given the sensitivity of pathogens to antibacterial drugs, the issue of treatment personalization appears to be of high importance [14][15].
Moreover, current treatments of chronic ME patients with corticosteroids, immunomodulators, and antibacterial agents are frequently insufficiently effective, and may have side effects, including additional disruption of their microbiome. In this regard, new therapies aimed not only at eliminating the symptoms of the disease, but also at restoring the disturbed microbiome should be found. Currently, the optimization of microbial eczema therapy by correcting the microbiome with probiotics has been studied, which may lead to improved treatment efficacy and more durable remission [16].
Bifida Ferment Lysate and Lactobacillus Ferment are the most commonly used ingredients in skin care products, classified as “probiotics”. They are fermentation products derived from Bifida and Lactobacillus bacteria, respectively. In contrast to systemic probiotics, external probiotics use lysates (decay products) of these bacteria. They regenerate the microbiome, strengthening the skin barrier, and have a soothing and antimicrobial effect [17]. In this context, topical probiotics should be included in the complex therapy of microbial eczema.
Consequently, the present study suggests optimizing the therapy of ME patients aimed at correcting the microbiome, which, in turn, could improve the efficacy of treatment of this pathology.
This study aims to evaluate the clinical efficacy of complex therapy of patients with microbial eczema with personalized selection of topical antibacterial agents taking into account the determination of antibiotic resistance by whole genome sequencing.
METHODS
Study design
The controlled randomized prospective study included 60 patients with chronic microbial eczema in the exacerbation stage, who underwent therapy, observation, and examination at the Department of Dermatovenerology of the Kuban State Medical University of the Ministry of Health of the Russian Federation.
Eligibility criteria
Inclusion criteria
Diagnosis of chronic microbial eczema (duration of disease at least 6 months); absence of other skin diseases; age between 18 and 70 years; absence of systemic and/or topical antibiotic therapy within the last month; signed informed consent.
Exclusion criteria
Presence of malignant neoplasms; history of chronic somatic diseases or diseases of neoplastic nature; diagnosis of acute microbial eczema; history of diseases associated with immunocompromised patients.
Removal criteria
Refusal of the patient to further participate in the study, refusal of dynamic follow-up.
Study conditions
The study was conducted at the Department of Dermatovenerology of the Kuban State Medical University of the Ministry of Health of the Russian Federation. The subjects were treated in accordance with the Federal Clinical Guidelines (FCG)2. The laboratory stage of the present study was performed at two sites: clinical diagnostic laboratory “CL Lab” (“CL Medicalgroup” LLC) in Krasnodar and genetic laboratory “Serbalab” (“Serbalab” LLC) in St. Petersburg.
Study duration
Recruitment and assignment of patients to groups, clinical study including laboratory stage was performed in the period from December 2023 to December 2024. Follow-up of the patients was carried out for 6 months.
Medical interventions
All the patients donated biomaterial (scrapings from the affected skin areas) for the laboratory stage of the study. The subjects were examined by a dermatovenerologist, anamnesis was collected, microscopic and bacteriologic examination of skin lesions was performed, and questionnaires were administered.
Patients in both groups were given standard therapy according to the FCG: antihistamines (cetirizine 10 mg/day in the evening for 14 days); systemic antibacterial agents (azithromycin 500 mg/day on the 1st day, then 250 mg once daily for 4 days); aniline dye solutions were applied externally (methylthioninium chloride 1% aqueous solution 2 times a day on the affected area for 7 days); detoxification drugs (sodium thiosulfate 30% solution intravenously 5.0–10.0 ml once a day for 10 days).
In the control group, topical glucocorticosteroids in combination with an antibacterial agent (betamethasone + gentamicin 0.1% + 0.1% topical cream 2 times a day on the affected skin areas for 14 days) were used.
Considering the study aim in the main group of patients, topical antibacterial agents were personalized based on antibiotic resistance data from WGS (betamethasone + fusidic acid, topical cream, once daily for 14 days from morning and fusidic acid 2%, topical cream, once daily for 14 days).
On the 21st day of therapy, an emollient with a probiotic component was added to the treatment regimen for all patients in both groups.
Study outcomes
Main study outcome
A study endpoint was considered the end of the treatment and follow-up period for 6 months. The efficacy of personalized use of topical antibiotics in complex therapy was evaluated according to the FCG3. Eczema Area and Severity Index (EASI) was used to analyze the severity of the course of chronic microbial eczema. The taxonomic microbial profile of skin lesions in patients of the compared groups was analyzed at different time points of observation.
Additional study outcomes
No additional outcomes are intended.
Methods for recording outcomes
The WGS method with interpretation was used to identify the taxonomic profile of the skin that affects the course of the disease. Biomaterial from patients with chronic ME was sampled from the affected skin areas before the start of therapy, on days 14, 21, and 6 months after treatment. For molecular genetic study, samples from lesions were collected by swabbing with disposable sterile cotton-tipped swabs. Swabs were previously wetted with sterile buffer solution and tightly rubbed for 20 seconds into the affected skin. The cotton swab samples were then stored in a tube filled with sterile buffer solution. Genomic DNA was extracted from each swab using a gel extraction kit. The further step involved lysis of bacterial cells by incubating samples with lysis buffer with homogenization by shaking with zirconium beads and performing a subsequent centrifugation. DNA genes from different regions were amplified using primer from variable regions 3–4, therefore broadening the taxonomic scope of the study. The result of WGS analysis was presented in conventional units (c. u.) of measurement. A conventional unit of measurement was equal to the number of reads of the nucleotide sequence of a particular microorganism among one hundred reads of all nucleotide sequences of a single patient biomaterial sample.
The EASI was used to evaluate the skin pathologic process [18][19]. The severity of the disease (mild, moderate, medium, severe, very severe) was evaluated in points and reflected clinical signs: erythema, edema, excoriations, lichenification (0.1–1.0 points for mild, 1.1–7.0 points for moderate, 7.1–21.0 points for medium, 21.1–50.0 points for severe, 50.1–72.0 points for very severe).
Randomization
For objectivity of the study, the envelope method was applied. Patients were randomly grouped separately for men (30 patients) and women (30 patients). Two packages were prepared, each with 30 opaque envelopes containing cards; 15 cards were labeled with the number “1” and the other 15 with the number “2”, meaning the number of the study group. Male patients picked envelopes with the group number from the male package while female patients picked envelopes from the female package, the number on the card determining which group the patient would be assigned to. This approach determined the equality of groups by sex thus guaranteeing random assignment of participants when groups were formed.
Data anonymization
The data were received and subsequently processed anonymously. A special key code was introduced for each patient without linking to personal data.
Statistical procedures
Principles behind sample size determination
The sample size was not determined in advance.
Statistical methods
In each patient of the main group (Group 1) and the control group (Group 2), the skin microbial contamination of different microorganisms was evaluated in conventional units. The primary microbiome data for statistical analysis were distributions of the occurrence of the studied pathological and nonpathological microorganisms measured for each taxon and for each patient in conventional units.
The primary data were statistically processed using the Statistica 12.0 software package (StatSoft, USA) and Microsoft Excel 2010 (Microsoft, USA).
Quantitative indicators for each patient in the main and the control groups were statistically described using the median (Me) and the first and third quartiles (Q1; Q3) written as (Me (Q1; Q3)) in case the hypothesis of normal distribution was rejected when tested by Kolmogorov—Smirnov and Shapiro—Wilk tests of normality. The nonparametric Mann—Whitney test was used to compare such indices in independent groups. Comparison of indicators of the same clinical group in different follow-up periods was performed using the nonparametric paired Wilcoxon test.
Quantitative parameters for each patient in Groups 1 and 2 were statistically described using the arithmetic mean (M) and standard deviation (SD) as a record (M ± SD) if the hypothesis of normal distribution was not rejected when tested by Kolmogorov—Smirnov and Shapiro—Wilk tests. The parametric Student’s t-test was used to compare such indicators in independent groups. Comparison of indicators of the same clinical group, albeit at different follow-up periods, was performed using the parametric paired Student’s t-test.
Structural or qualitative indicators between different clinical groups were compared using Pearson’s Chi-square test or Fisher’s exact test for small numbers of sample members. Differences were considered statistically significant at an error level of p < 0.05.
RESULTS
Sampling
The sampling procedure and the study’s design are presented in the block diagram (Fig. 1). The subjects of the two groups donated biomaterial (scraping from the affected skin areas) for laboratory tests. The overall number of patients was formed by including patients with chronic microbial eczema in accordance with the inclusion criteria for patients in the study. Two groups were randomly formed: main (Group 1) and control (Group 2). Patients in the control group (n = 30) received standard therapy according to the FCG. The main group (n = 30) also received conventional therapy; however, topical antibacterial agents were personalized based on antibiotic resistance data from WGS.
Characteristics of the study sample (groups)
Patients randomly selected into the study groups were comparable in various characteristics before treatment. Table 1 shows the anthropometric characteristics.
In both groups, 15 women (15/30, 50.00%) and 15 men (15/30, 50.00%) were under observation, i.e., groups were completely consistent in terms of sex. The mean age of patients in Group 1 was 37.50 ± 13.71 years and in Group 2 was 41.43 ± 16.14 years, with no statistical significance of differences in mean age (p = 0.313), meaning that the groups were comparable before treatment. The values of mean height in Groups 1 and 2 were also comparable. In Group 1, the mean height was 171 ± 9 cm, while in Group 2 it was 170 ± 8 cm, the differences were not significant (p = 0.832). In terms of patient weight, the mean values differed. Namely, in Group 1 the mean weight was 70.10 ± 14.20 kg and in Group 2 it was 67.50 ± 13.80 kg, although the differences were not found to be statistically significant (p = 0.475).
Similarly to the above-mentioned anthropometric characteristics, the body mass index (BMI) indicators differed for the mean values of the groups, though not statistically significant (p = 0.35). Therefore, the groups were comparable in terms of sex, age, height, weight, and BMI.
The comparability of the groups can be considered by examining the microbial profile of patients in these groups (Table 2).
In pre-treatment skin microbiota of patients in both groups, P. aeruginosa and E. coli were more prominent with median values of 7.04 (5.53; 8.43) c. u. and 9.39 (8.34; 9.75) c. u., respectively, in Group 1 among the total number of isolated bacteria in the rash area. Similarly, P. aeruginosa and E. coli exhibited median values of 7.04 (5.52; 8.42) c. u. and 9.49 (8.34; 9.84) c. u., respectively, in Group 2 among the total number of isolated bacteria in the rash area. Comparison of median values between groups showed no statistically significant difference for both P. aeruginosa (p = 0.929) and E. coli (p = 0.717). In addition, S. aureus (median and quartiles were 2.05 (1.86; 2.44) c. u.) and K. pneumonia (median and quartiles were 4.44 (4.01 5.95) c. u.) were sequenced on skin sections from patients in Group 1 (main). S. aureus (median and quartiles were 2.04 (1.84; 2.44) c. u.) and K. pneumonia (median and quartiles were 4.43 (4.01 5.94) c. u.) were also sequenced in Group 2 (control) patients. Differences were not significant (p = 0.988 for S. aureus and p = 0.994 for K. pneumonia). In Group 1 patients, the microbial pathogenic community included C. difficile (1.97 (1.51; 2.14) c. u.), M. tuberculosis (0.94 (0.79; 1.08) c. u.). Similarly in Group 2 patients were detected C. difficile (1.96 (1.50; 2.13) c. u.), M. tuberculosis (0.93 (0.78; 1.07) c. u.). For these two microorganisms, the differences between the groups were not significant (p > 0.05). Small values of indices were observed in both groups and for functional bacteria, which also showed no significant differences (Table 2).
Thus, the groups are comparable in terms of microbiome profile.
Then, consider whether the groups are comparable in some clinical parameters for the patients in the studied groups (Table 3).
In the main group before treatment, 17/30 (56.67%) patients had moderate microbial eczema and 13/30 (43.33%) patients had severe eczema according to the EASI values; the median value of this index was 17.94 (10.51; 29.10). In the control group, 19/30 patients (63.33%) had a medium degree of severity of ME, and 11/30 patients (36.67%) had a severe course of the disease. The difference in the proportion of patients with moderate and severe disease in the compared groups was not statistically significant (p = 0.599). The median index value in Group 2 was 18.03 (11.32; 29.59) points, which was comparable to the index value in the main group of patients (p > 0.05).
The duration of the disease ranged from 6 to 36 months.
Hence, such clinical indicators as EASI index, duration of disease, the proportion of patients with different degrees of disease severity before the start of treatment in Groups 1 and 2 were comparable.
Consequently, in terms of anthropometric characteristics, microbiome profile, and clinical parameters, both groups of patients were close in their values before the intervention, i.e., the different treatments of the present study.
Main study results
Before treatment, antibiotic resistance of bacteria was determined in the main group of patients. A total of 21 names were identified; the shares of different species among all identified species are shown in Figure 2.
In the main group, 11/30 (36.67%) patients had no resistance, 9/30 patients had resistance only to aminoglycosides (30.00%), 4/30 (13.33%) only to macrolides, 1/30 (3.33%) only to diaminopyrimidines, 1/30 (3.33%) to oxazolidinones, 2/30 (6.67%) to fluoroquinolones. In 2/30 (6.67%) patients, resistance to two aminoglycosides was established simultaneously, each to diaminopyrimidines and macrolides, and to aminoglycosides.
A topical antibacterial agent with fusidic acid, which has a bactericidal effect, was chosen for the study in the main group of patients. It suppresses protein synthesis by inhibiting the factor necessary for translocation of protein subunits and elongation of the peptide chain, which further leads to the death of the pathogen.
Let us consider the changes in the microbial prevalence rate at different follow-up times during treatment for the patients of the main group (Group 1) and the control group (Group 2).
In all patients before treatment, opportunistic bacteria with medium and high values of microbial index in conventional units were identified, such as S. aureus, P. aeruginosa, E. coli, K. pneumoniae, C. difficile, and M. tuberculosis. Functional bacteria with very low index values were also sequenced including Lactobacillus, Bifidobacterium, and S. epidermidis.
On the day 14 of treatment, the index for S. aureus on the skin in Group 1 patients decreased compared to the situation before treatment to 1.82 (1.70; 2.00) c. u., while in Group 2 this index was greater and amounted to 2.02 (1.84; 2.39) c. u., indicating a statistically significant difference (p = 0.004). Meanwhile, the P. aeruginosa contamination decreased to 4.84 (2.99; 5.94) c. u. in the main group, while in the control group the value was higher and corresponded to 5.31 (3.83; 6.52) c. u., though not statistically significant (p = 0.246 compared to the results of the main group for this follow-up period). Group 1 patients had a lower index for E. coli on day 14 of treatment than before treatment and lower than in Group 2. Thus, the index for Group 1 was 5.52 (4.90; 5.73) c. u., while in Group 2 it was 8.49 (7.58; 8.94) c. u. at p < 0.001, i.e., the difference in medians by a value equal to 2.97 c. u. was statistically significant. No significant difference in the follow-up period of 14 days for K. pneumonia microorganisms was found for Groups 1 and 2 (p = 0.877). Similarly, no differences of the same indicator were found for Groups 1 and 2 and for microorganisms with taxons C. difficile (p = 0.751) and M. tuberculosis (p = 0.912).
Among functional bacteria, a slightly higher value of the index for Bifidobacterium and Lactobacillus was observed on the affected skin in the patients of the main group on the 14th day of treatment compared to the values before treatment. However, comparison of the values for Groups 1 and 2 on day 14 from the beginning of treatment for functional bacteria did not give a statistically significant difference in the medians obtained in the main group and the control group (Table 4).
On the 21st day of treatment the patients also showed negative dynamics of pathogens contamination on the skin areas. Whereas on the 14th day of treatment, only two pathogens were differentiated, on the 21st day there was a difference in changes for three pathogens and one functional bacterium. The lower value of the index in the main group compared to Group 2 was for E. coli (p < 0.001), K. pneumonia (p = 0.041), M. tuberculosis (p = 0.017), and higher value for Bifidobacterium (p < 0.001).
Additionally, a lower number of reads out of 100 reads in the isolated sample of patient’s biomaterial was noted for S. aureus, equal to 0.99 (0.81; 1.07) c. u. in the main group compared to Group 2 with a median of 1.05 (0.80; 1.86) c. u. (p = 0.050).
Non-significant changes in rates between the two groups of comparison at day 21 of treatment were observed for the remaining bacteria, P. aeruginosa, C. difficile, and the functional bacteria, S. epidermidis, Lactobacillus.
After 6 months of treatment, the medians of prevalence rates for 7 out of 9 observed bacteria were found to be statistically significantly different between the main group and the control group (Table 6). The median values for E. coli and S. aureus in Groups 1 and 2 were similar. The median values for P. aeruginosa, K. pneumonia, C. difficile, and M. tuberculosis in Group 1 were significantly lower than in Group 2, while the median values for S. epidermidis, Lactobacillus, Bifidobacterium in Group 1 were significantly higher than in Group 2.
After 14 days, the prevalence rate of two pathogenic bacteria E. coli and S. aureus was significantly lower in the main group compared to the control group. After 21 days, the prevalence rate was significantly lower already for as many as four pathogenic bacteria and higher for one functional bacterium.
By 6 months, the values for seven bacteria (four from the pathologic pool and three from the functional pool) in Group 1 were closer to the values for recovery than the values for the same bacteria in Group 2.
After 6 months of treatment, a statistically significant decrease of P. aeruginosa in the foci of lesions in the patients of the main group to 1.16 (1.02; 1.67) c. u. (p < 0.001 in comparison with the values before the start of treatment), while in the control group the values were 0.46 c. u. higher (1.62 (1.22; 1.98) c. u.) (p < 0.001 in comparison with the values in the main group) (Table 6).
Furthermore, in the main group, K. pneumonia and C. difficile had an index equal to 1.05 (0.97; 1.43) c. u. and 0.09 (0.06; 0.32) c. u., respectively (p < 0.001 as compared to the values before the start of treatment for both K. pneumonia and C. difficile), while in the control group the index was 0.94 c. u. higher (1.99 (1.75; 2.09) c. u.) at p < 0.001 compared to the main group, and 0.21 c. u. higher (0.30 (0.13; 0.79) c. u.), respectively, at p = 0.003 compared to the main group.
In the lesion foci of patients with personalized topical antibiotic, sites of S. epidermidis metagenome were sequenced among functional species of the genus Staphylococcus (0.30 (0.24; 0.33) c. u.). The control group had the index 0.13 (0.17 (0.15; 0.27) c. u.) less (p = 0.001 compared to the result of the main group).
The index in representatives of functional flora, Bifidobacterium and Lactobacillus, was increased compared to the values before treatment by 2.31 and 2.10 times, respectively (p < 0.001 with the start of treatment) in Group 1, while in Group 2 they were increased only by 1.80 and 1.67 times, respectively (p < 0.001 compared to the main group) (Tables 6 and 7). Since both groups of patients were treated, the non-parametric paired (for dependent samples) Wilcoxon test was used to determine that a statistically significant change in prevalence (with high statistical significance of p < 0.01) occurred for all bacteria and for all follow-up periods in both groups compared to pre-treatment values. These changes tended to decrease for pathogenic bacteria and to increase for functional bacteria. The ratio of the values of indicators before treatment and 6 months after the start of treatment is shown in Table 7. For example, the ratios of indicators for the control group for most bacteria (excluding E. coli and S. aureus) are significantly less than the similar ratios for the main group. The decrease in the prevalence of E. coli and S. aureus is approximately the same: 5.49 times less in Group 1 and 5.18 times less in Group 2 (Table 7).
The values of microbial parameters were compared between groups using the nonparametric Mann—Whitney criterion (for independent samples) to analyze the treatment efficacy. The grouping of statistically significant changes at different follow-up periods and for different microorganisms revealed three types of response to different methods of treatment (Table 8).
Type 1 of bacterial behavior during treatment in both groups corresponded to E. coli and S. aureus bacteria, in which the decrease in microbial parameter by 14 days from the start of treatment was greater in the group of patients with personalized topical antibiotic than in Group 2. The same effect was observed at 21 days from the start of treatment. By a follow-up period of 6 months from the start of treatment, the values for these microorganisms were approximately the same, as the differences were not significant (p = 0.290 and p = 0.176 between Groups 1 and 2). The median values at different follow-up times in both groups for Type 1 of behavior using E. coli as an example are shown in Figure 3.
Type 2 of bacterial response to treatment in both groups corresponded to K. pneumonia and M. tuberculosis bacteria, in which only by 21 days from the start of treatment the reduction in microbial parameter in the group of patients with personalized topical antibiotic was significantly greater than in the control group. By the period of 6 months from the start of treatment the parameters for these microorganisms were significantly different. In the main group it was less than in Group 2 (for K. pneumonia p < 0.001 and for M. tuberculosis p = 0.018). The median values at different follow-up times in both groups for Type 2 of behavior exemplified by K. pneumonia are shown in Figure 4.
Type 3 of bacterial behavior during treatment in both groups corresponded to P. aeruginosa and C. difficile, in which only by 6 months from the start of treatment the difference in microbial parameter with lower median values in Group 1 compared to Group 2 was statistically significant (for P. aeruginosa p = 0.011 and for C. difficile p = 0.003). The values of medians at different follow-up periods in both groups for Type 3 of behavior on the example of P. aeruginosa are shown in Figure 5.
The behavior of functional bacteria appears to be similar to Type 2 (for Bifidobacterium) and Type 3 (for S. epidermidis and Lactobacillus).
The EASI index was changing in the following way: before the treatment the median values for the groups nearly coincided (p = 0.706) (Table 9); in the main group on the 14th day of treatment the median value of the EASI index amounted to 3.05 (1.64; 5.33) points, while in the control group it was 5.53 (4.14; 9.88) points (p < 0.001 compared to the values in the main group); on day 21 of treatment, the median value of the EASI index was 1.00 (0.53; 1.25) points in the main group and 1.84 (0.77; 4.22) points in the control group (p = 0.006 compared to the main group).
By the end of follow-up (month 6), the EASI index in the main group decreased from the beginning of treatment and amounted to 0.70 (0.24; 1.00) points versus 1.00 (0.77; 4.22) in the control group at p < 0.001. The median value for the main group decreased 25.63 times compared to the value before treatment. The median EASI was only 18.03 times lower in the control group, proving the greater efficacy of treatment with personalized topical antibiotics.
The share of patients with relapse and different degrees of severity of residual disease is another evidence of the treatment efficacy in the main group. The mild degree of the disease course was observed in 25/30 (83.33%) people, moderate in 3/30 (10.00%), and medium in 2/30 (6.67%). Two patients in the main group had a relapse of the disease. They complained of erythema and edema. In the control group, mild course of microbial eczema was found in 16/30 (53.33%) patients, moderate in 9/30 (30.00%), and medium in 5/30 (16.67%). Five patients relapsed with complaints of erythema, single papular elements, lichenification, and itching. The statistically significant difference in the shares of patients with different degrees of disease severity in the compared groups was examined using crosstabulation analysis and Fisher’s exact criterion in the form of a quadratic table (Table 10).
The share of patients with a mild degree of the disease in the main group, equal to 83.33%, statistically significantly exceeds the similar share of patients in the control group, equal to 53.33%, indicating greater treatment efficacy in the main group.
Additional study results
No additional results were obtained during the course of the study.
Adverse events
No adverse events have been reported.
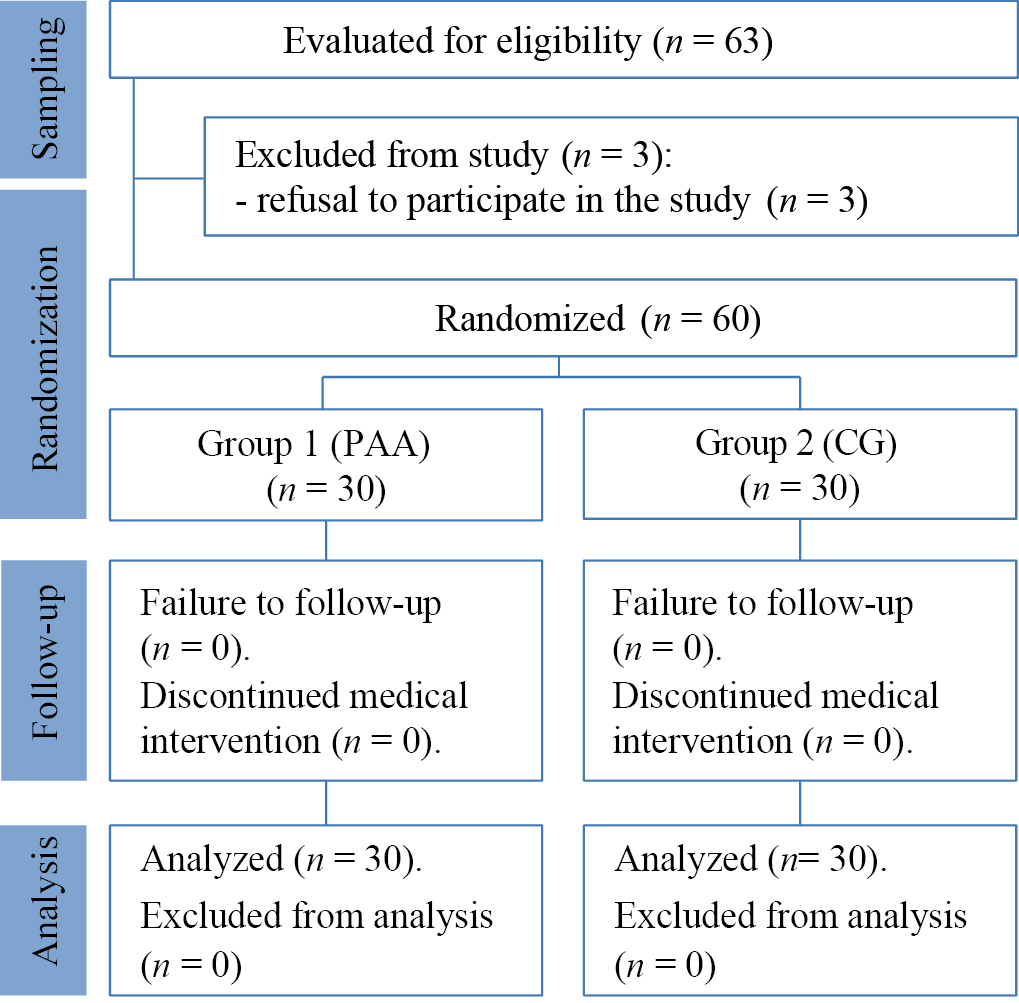
Fig. 1. Block diagram of the study design
Note: the block diagram was created by the authors (according to STROBE recommendations). Abbreviations: CG—therapy according to clinical guidelines; PAA—therapy with personalized selection of antimicrobial agents.
Рис. 1. Блок-схема дизайна исследования
Примечание: блок-схема выполнена авторами (согласно рекомендациям STROBE). Сокращения: CG — терапия согласно клиническим рекомендациям; PAA — терапия с персонализированным подбором антимикробного средства.
Table 1. Anthropometric characteristics of the studied groups of patients
Таблица 1. Антропометрические характеристики исследуемых групп больных
|
Characteristic |
Group 1 (n = 30) |
Group 2 (n = 30) |
Statistical significance p |
|
Sex, (n)male / (n)female |
15/15 |
15/15 |
|
|
Age, years |
37.50 ± 13.71 |
41.43 ± 16.14 |
0.313 |
|
Height, cm |
171 ± 9 |
170 ± 88 |
0.832 |
|
Weight, kg |
70.1 ± 14.2 |
67.5 ± 13.8 |
0.475 |
|
BMI, kg/m2 |
23.6 ± 2.9 |
22.9 ± 3.1 |
0.354 |
Note: the table was compiled by the authors. Abbreviation: BMI—body mass index.
Примечание: таблица составлена авторами. Сокращение: BMI — индекс массы тела.
Table 2. Microbiome profile (Me (Q1; Q3)) in patients in the studied groups before treatment
Таблица 2. Микробиомный профиль в виде Ме (Q1; Q3) у больных в исследуемых группах до лечения
|
Microbiome |
Group 1 (n = 30) |
Group 2 (n = 30) |
Statistical significance p |
|
E. coli |
9.39 (8.34; 9.75) |
9.49 (8.34; 9.84) |
0.717 |
|
P. aeruginosa |
7.04 (5.53; 8.43) |
7.04 (5.52; 8.42) |
0.929 |
|
K. pneumonia |
4.44 (4.01; 5.95) |
4.43 (4.01; 5.94) |
0.994 |
|
S. aureus |
2.05 (1.86; 2.44) |
2.04 (1.84; 2.44) |
0.988 |
|
C. difficile |
1.97 (1.51; 2.14) |
1.96 (1.50; 2.13) |
0.994 |
|
M. tuberculosis |
0.94 (0.79; 1.08) |
0.93 (0.78; 1.07) |
0.993 |
|
S. epidermidis |
0.03 (0.02; 0.05) |
0.02 (0.01; 0.05) |
0.631 |
|
Lactobacillus |
1.01 (0.88; 1.27) |
1.02 (0.86; 1.27) |
0.906 |
|
Bifidobacterium |
0.35 (0.32; 0.39) |
0.35 (0.30; 0.38) |
0.848 |
Note: the table was compiled by the authors.
Примечание: таблица составлена авторами.
Table 3. Clinical parameters in patients in the studied groups
Таблица 3. Клинические показатели у больных в исследуемых группах
|
Parameter |
Group 1 (n = 30) |
Group 2 (n = 30) |
Statistical significance р |
|
EASI, point |
17.94 (10.51; 29.10) |
18.03 (11.32; 29.59) |
0.706 |
|
Disease duration, months |
18.5 (9.5; 29.3) |
18.0 (12.0; 29.8) |
0.871 |
|
Disease severity |
|||
|
Medium, number/share |
17/0.57 |
19/0.63 |
0.599 |
|
Severe, number/share |
13/0.43 |
11/0.37 |
0.599 |
Note: the table was compiled by the authors. Abbreviation: EASI—Eczema Area and Severity Index.
Примечание: таблица составлена авторами. Сокращение: EASI (Eczema Area and Severity Index) — индекс площади поражения и тяжести атопического дерматита.
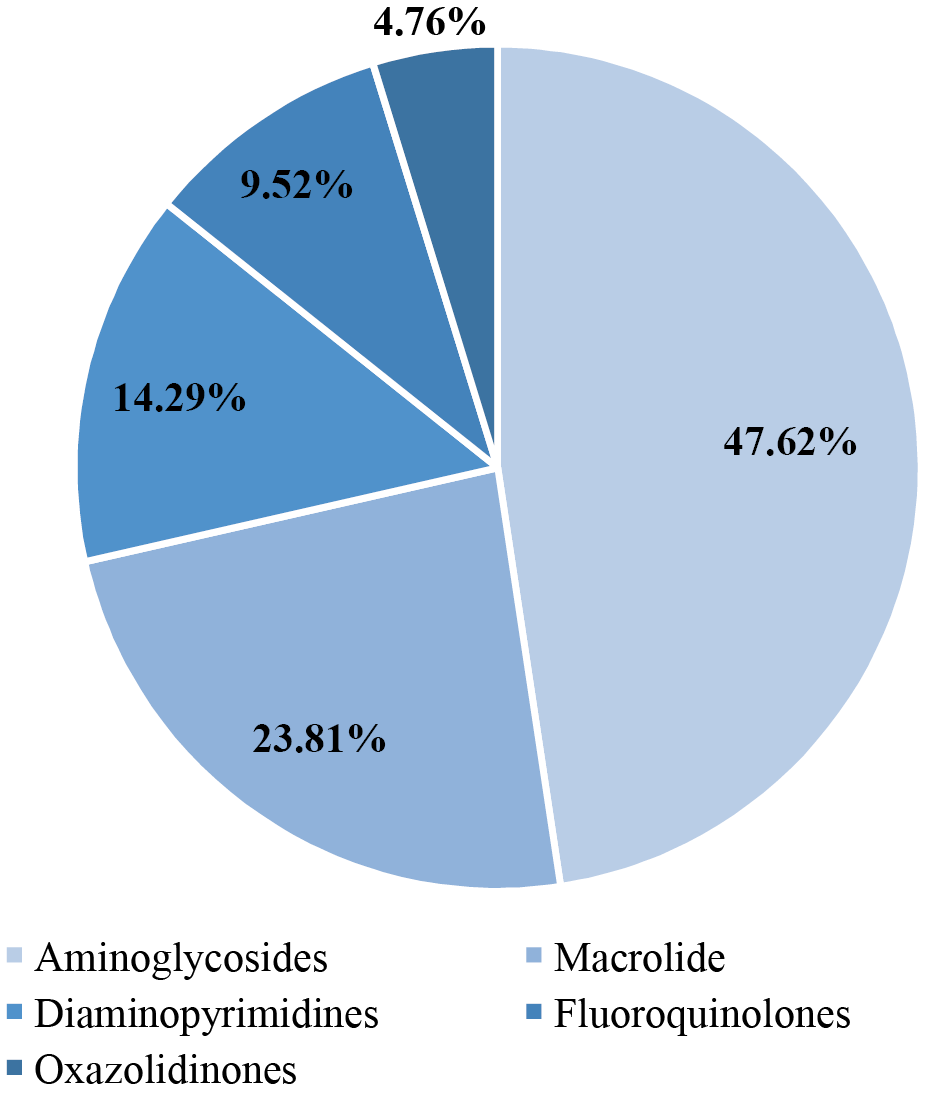
Fig. 2. Groups of antibiotics to which Staphylococcus aureus is resistant
Note: the figure was created by the authors.
Рис. 2. Группы антибиотиков, к которым резистентна Staphylococcus aureus
Примечание: рисунок выполнен авторами.
Table 4. Microbiome profile (Me (Q1; Q3)) in patients in the study groups on day 14
Таблица 4. Микробиомный профиль в виде Ме (Q1; Q3) у больных в исследуемых группах на 14-й день
|
Microbiome |
Group 1 (n = 30) |
Group 2 (n = 30) |
Statistical significance р* |
|
E. coli |
5.52 (4.90; 5.73) |
8.61 (7.58; 8.94) |
< 0.001 |
|
P. aeruginosa |
4.84 (2.99; 5.94) |
5.31 (3.83; 6.51) |
0.245 |
|
K. pneumonia |
4.44 (4.00; 5.72) |
4.43 (4.01; 5.94) |
0.876 |
|
S. aureus |
1.82 (1.65; 1.98) |
2.02 (1.84; 2.39) |
0.003 |
|
C. difficile |
1.95 (1.51; 2.10) |
1.96 (1.50; 2.14) |
0.750 |
|
M. tuberculosis |
0.93 (0.78; 1.06) |
0.93 (0.79; 1.08) |
0.911 |
|
S. epidermidis |
0.04 (0.03; 0.05) |
0.04 (0.03; 0.05) |
0.773 |
|
Lactobacillus |
1.11 (0.95; 1.40) |
1.02 (0.88; 1.27) |
0.125 |
|
Bifidobacterium |
0.39 (0.34; 0.43) |
0.38 (0.33; 0.41) |
0.399 |
Note: the table was compiled by the authors; p* — by the Mann—Whitney criterion.
Примечание: таблица составлена авторами; р* — по критерию Манна — Уитни.
Table 5. Microbiome profile (Me (Q1; Q3) in patients in the studied groups on day 21 of treatment
Таблица 5. Микробиомный профиль в виде Ме (Q1; Q3) у больных в исследуемых группах на 21-й день лечения
|
Microbiome |
Group 1 (n = 30) |
Group 2 (n = 30) |
Statistical significance р* |
|
E. coli |
4.60 (4.08; 4.77) |
6.32 (5.55; 6.53) |
< 0.001 |
|
P. aeruginosa |
4.50 (2.98; 5.49) |
5.00 (3.79; 5.79) |
0.442 |
|
K. pneumonia |
3.42 (3.02; 4.05) |
3.94 (3.22; 4.93) |
0.041 |
|
S. aureus |
0.99 (0.81; 1.07) |
1.05 (0.80; 1.86) |
0.050 |
|
C. difficile |
1.53 (1.11; 1.96) |
1.38 (1.01; 1.97) |
0.853 |
|
M. tuberculosis |
0.63 (0.51; 0.75) |
0.72 (0.62; 0.83) |
0.017 |
|
S. epidermidis |
0.10 (0.08; 0.11) |
0.09 (0.06; 0.12) |
0.214 |
|
Lactobacillus |
1.13 (0.96; 1.42) |
1.03 (0.89; 1.27) |
0.131 |
|
Bifidobacterium |
0.55 (0.47; 0.59) |
0.40 (0.35; 0.43) |
< 0.001 |
Notes: the table was compiled by the authors; p* — by the Mann—Whitney criterion.
Примечания: таблица составлена авторами; р* — по критерию Манна — Уитни.
Table 6. Microbiome profile (Me (Q1; Q3) in patients in the studied groups on the 6th month of treatment
Таблица 6. Микробиомный профиль в виде Ме (Q1; Q3) у больных в исследуемых группах на 6-й месяц лечения
|
Microbiome |
Group 1 (n = 30) |
Group 2 (n = 30) |
Statistical significance р* |
|
E. coli |
1.71 (1.04; 2.17) |
1.83 (1.23; 2.41) |
0.290 |
|
P. aeruginosa |
1.16 (1.02; 1.67) |
1.62 (1.22; 1.98) |
0.011 |
|
K. pneumonia |
1.05 (0.97; 1.43) |
1.99 (1.75; 2.09) |
< 0.001 |
|
S. aureus |
0.65 (0.46; 0.80) |
0.69 (0.61; 0.90) |
0.176 |
|
C. difficile |
0.09 (0.06; 0.32) |
0.30 (0.13; 0.79) |
0.003 |
|
M. tuberculosis |
0.38 (0.27; 0.50) |
0.50 (0.38; 0.60) |
0.018 |
|
S. epidermidis |
0.30 (0.24; 0.33) |
0.17 (0.15; 0.27) |
0.001 |
|
Lactobacillus |
2.12 (1.83; 2.67) |
1.70 (1.46; 2.10) |
< 0.001 |
|
Bifidobacterium |
0.81 (0.72; 0.89) |
0.63 (0.57; 0.68) |
< 0.001 |
Note: the table was compiled by the authors; p* — by the Mann—Whitney criterion.
Примечание: таблица составлена авторами; р* — по критерию Манна — Уитни.
Table 7. Median values of bacterial prevalence rates before treatment and 6 months after the start of treatment in the studied groups of patients
Таблица 7. Отношения медианных значений показателей распространенности бактерий до лечения и через 6 месяцев после начала лечения в исследуемых группах больных
|
Microbiome |
Group 1 (n = 30) |
Group 2 (n = 30) |
Statistical significance р* |
|
E. coli |
5.49 |
5.18 |
0.290 |
|
P. aeruginosa |
6.07 |
4.35 |
0.011 |
|
K. pneumonia |
4.23 |
2.23 |
< 0.001 |
|
S. aureus |
3.15 |
2.96 |
0.176 |
|
C. difficile |
21.89 |
6.53 |
0.003 |
|
M. tuberculosis |
2.47 |
1.86 |
0.018 |
|
S. epidermidis |
10.00 |
8.50 |
0.001 |
|
Lactobacillus |
2.10 |
1.67 |
< 0.001 |
|
Bifidobacterium |
2.31 |
1.80 |
< 0.001 |
Notes: the table was compiled by the authors; p* — by the Mann—Whitney criterion.
Примечания: таблица составлена авторами; р* — по критерию Манна — Уитни.
Table 8. Statistical significance levels of differences in median values of microbial parameters of patients by type of response to therapy for different periods of observation in the studied groups
Таблица 8. Значения уровней статистической значимости различий медианных значений микробных показателей больных по типу ответа на терапию для разных сроков наблюдения в исследуемых группах
|
Microbiome |
Before treatment |
After 14 days |
After 21 days |
After 6 months |
|
Type 1 of bacterial behavior |
||||
|
E. coli |
0.717 |
р < 0.001 |
р < 0.001 |
p = 0.290 |
|
S. aureus |
0.988 |
p = 0.003 |
p = 0.050 |
p = 0.176 |
|
Type 2 of bacterial behavior |
||||
|
K. pneumonia |
0.994 |
p = 0.876 |
p = 0.041 |
р < 0.001 |
|
M. tuberculosis |
0.993 |
p = 0.911 |
p = 0.017 |
p = 0.018 |
|
Type 3 of bacterial behavior |
||||
|
P. aeruginosa |
0.929 |
p = 0.245 |
p = 0.442 |
p = 0.011 |
|
C. difficile |
0.994 |
p = 0.750 |
p = 0.853 |
p = 0.003 |
|
Functional bacteria |
||||
|
S. epidermidis |
0.631 |
p = 0.773 |
p = 0.214 |
p = 0.001 |
|
Lactobacillus |
0.906 |
p = 0.125 |
p = 0.131 |
р < 0.001 |
|
Bifidobacterium |
0.848 |
p = 0.399 |
р < 0.001 |
р < 0.001 |
Note: the table was compiled by the authors.
Примечание: таблица составлена авторами.
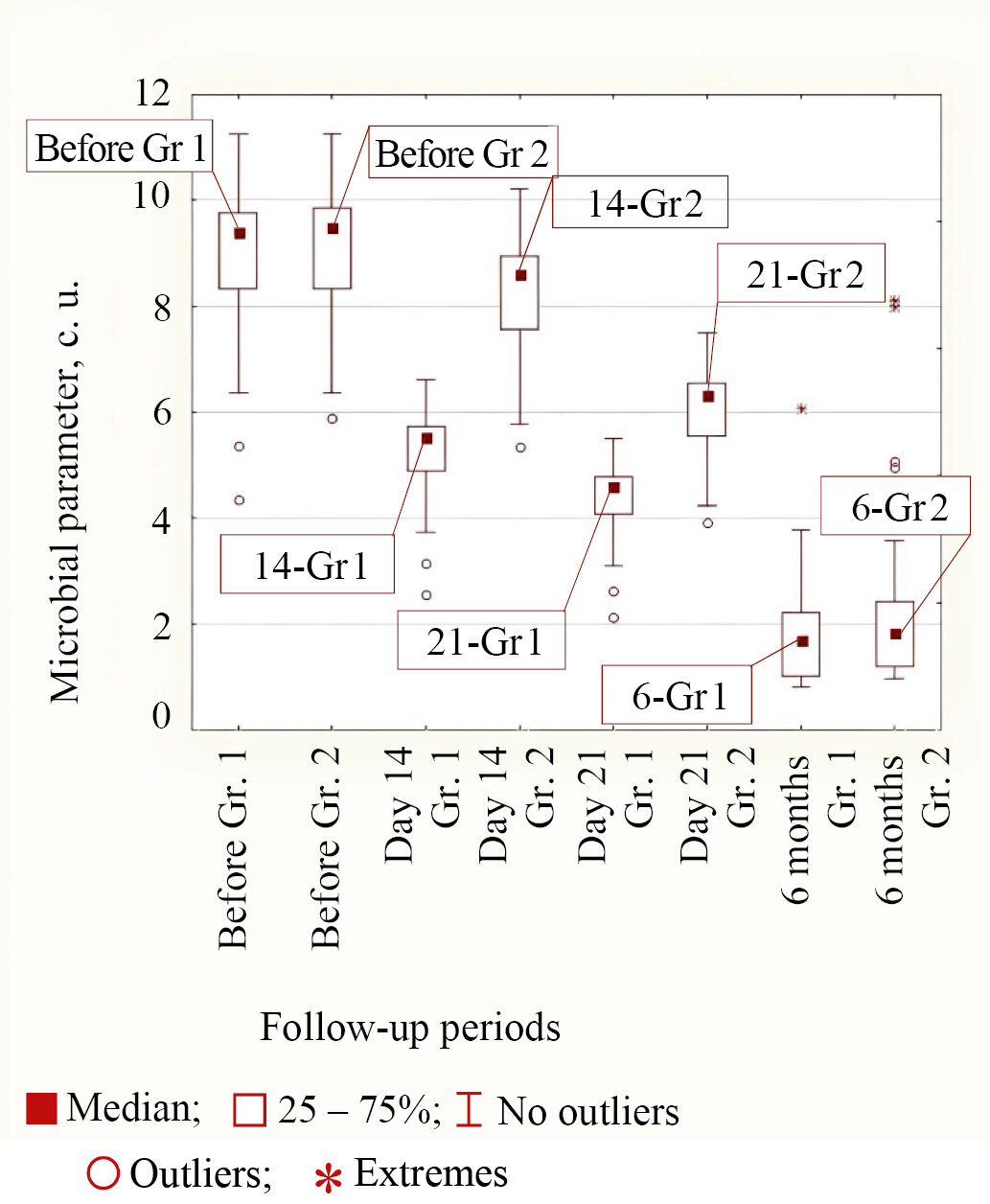
Fig. 3. Medians of microbial parameter values for E. coli bacteria in groups (Group 1 is the main group, Group 2 is the control group) for different observation periods
Note: the figure was made by the authors. Abbreviation: Gr—group.
Рис. 3. Значения медиан микробного показателя для бактерии E. coli в группах (группа 1 — основная группа, группа 2 — группа сравнения) для различных сроков наблюдения
Примечание: рисунок выполнен авторами. Сокращение: Gr — группа.
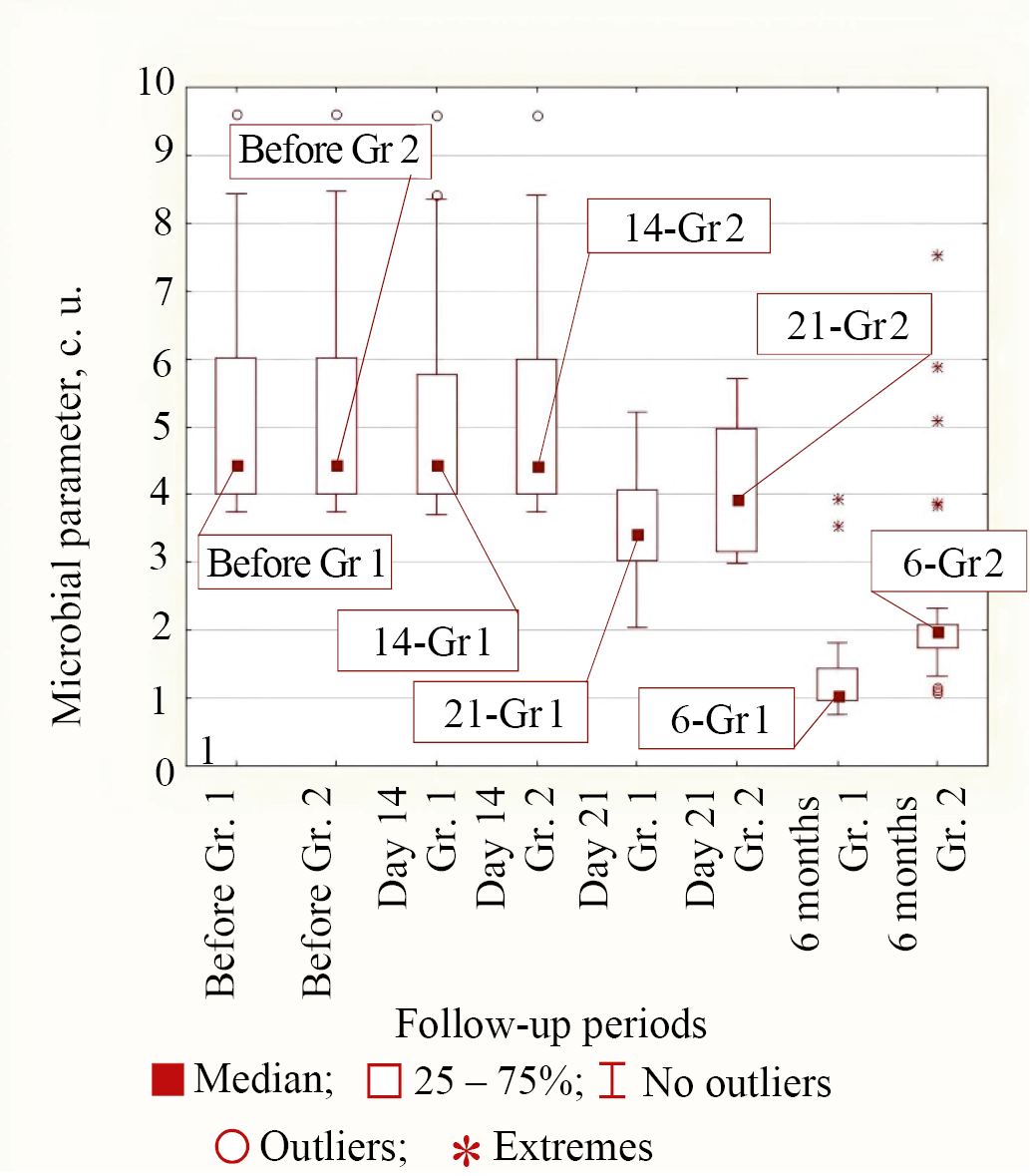
Fig. 4. Medians of microbial parameter values for K. pneumonia bacteria in groups (Group 1 is the main group, Group 2 is the control group) for different observation periods
Note: the figure was made by the authors. Abbreviation: Gr—group.
Рис. 4. Значения медиан микробного показателя для бактерии K. pneumonia в группах (группа 1 — основная группа, группа 2 — группа сравнения) для различных сроков наблюдения
Примечание: рисунок выполнен авторами. Сокращение: Gr — группа.
Table 9. Medians of the EASI index for different observation periods in the compared groups of patients
Таблица 9. Значения медиан индекса EASI для различных сроков наблюдения в сравниваемых группах больных
|
Observation period |
Group 1 (n = 30) |
Group 2 (n = 30) |
Statistical significance р* |
|
Before treatment |
17.94 (10.51; 29.10) |
18.03 (11.32; 29.59) |
0.706 |
|
After 14 days |
3.05 (1.64; 5.33) |
5.53 (4.14; 9.88) |
< 0.001 |
|
After 21 days |
1.00 (0.53; 1.25) |
1.84 (0.95; 4.01) |
0.006 |
|
After 6 months |
0.70 (0.24; 1.00) |
1.00 (0.77; 4.22) |
< 0.001 |
Notes: the table was compiled by the authors; p* — by the Mann—Whitney criterion. Abbreviation: EASI—Eczema Area and Severity Index
Примечания: таблица составлена авторами; р* — по критерию Манна — Уитни. Сокращение: EASI (Eczema Area and Severity Index) — индекс площади поражения и тяжести атопического дерматита.
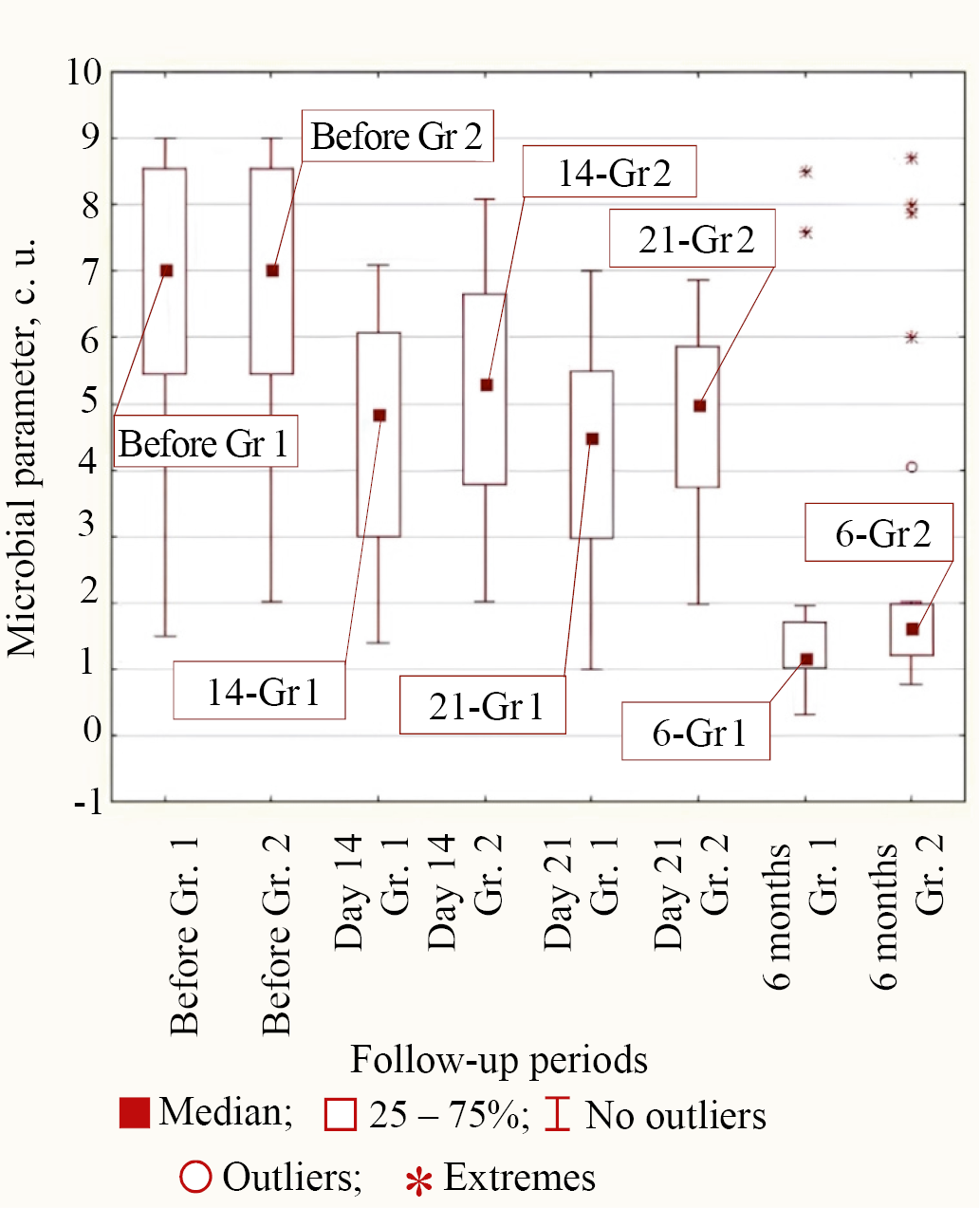
Fig. 5. Medians of microbial parameter values for P. aeruginosa bacteria in groups (Group 1 is the main group, Group 2 is the control group) for different observation periods
Note: the figure was made by the authors. Abbreviation: Gr—group
Рис. 5. Значения медиан микробного показателя для бактерии P. aeruginosa в группах (группа 1 — основная группа, группа 2 — группа сравнения) для различных сроков наблюдения
Примечание: рисунок выполнен авторами. Сокращение: Gr — группа.
Table 10. Number of patients and their shares with different severity of residual disease in the compared groups
Таблица 10. Количество больных и их доли с различными степенями тяжести остаточного заболевания в группах сравнения
|
Disease severity |
Group 1 (n = 30) |
Group 2 (n = 30) |
Statistical significance р* |
|
Mild, abs/rel% |
25/83.33 |
16/53.33 |
0.015 |
|
Moderate, abs/rel% |
3/10.00 |
9/30.00 |
0.063 |
|
Medium, abs/rel% |
2/6.67 |
5/16.67 |
0.263 |
Notes: the table was compiled by the authors; p* — by Chi-square test or Fisher’s exact test
Примечания: таблица составлена авторами; р* — по критерию Хи-квадрат или по точному критерию Фишера.
DISCUSSION
Study limitations
Relatively low statistical power due to the limited number of patients may be a factor potentially limiting the study.
Generalizability/extrapolation
The results of this study may apply to other clinical and experimental settings. For example, when other topical antibacterial agents other than those used in this study are used, WGS can exhibit a similar tendency to alter the microbiome of affected skin areas.
Summary of the main study result
The study analyzed the composition of the microbiome of the affected skin areas of ME patients. Before the start of therapy, the WGS revealed that the skin was mostly inhabited by opportunistic bacteria, and the number of functional bacteria was limited. The proposed complex therapy with the use of FCG and personalized topical antibacterial agents showed high efficiency in the therapy of chronic microbial eczema, which is confirmed by the decrease in the share of pathogens in the main group to a greater extent than in the control group, and by the reduced number of disease relapses.
Interpretation of the main study result
At the end of treatment, the comparative analysis of the treatment results of the patients of both groups showed a higher manifestation of dysbiotic changes on the studied skin areas in the control group.
Antibiotic resistance of bacteria on the skin has been previously studied by other researchers using the method of serial dilutions in agar or the diffusion method using antibiotic-impregnated disks [20]. Some experts have noted that the greatest therapeutic efficacy among topical antibiotics is indeed observed for fusidic acid and mupirocin, which is consistent with the study [21–25]. Other researchers in their works prefer a combination of two or three topical antibiotics to which resistance of Gram-positive and Gram-negative bacteria develops relatively slowly [26–28]. However, the method of personalized antibiotic therapy of skin diseases taking into account WGS data has not been previously investigated.
The study conducted using the WGS method confirms the resistance of S. aureus on the skin to a number of antibacterial drugs, also providing an opportunity for personalized selection of topical agents. The treatment performed in the main group of patients decreased the number of relapses by 2.5 times.
Analysis of the dynamics of the skin pathological process revealed an earlier regression and a distinct decrease in the severity of the eczematous process in patients whose treatment regimen included a personalized topical antibiotic. Moreover, already on day 14, the proportion of representatives of functional flora in patients of this group became greater (1.1 times at p < 0.001), which was not characteristic of patients with standard therapy.
The results obtained during WGS indicate that the complex therapy proposed in the main group with the use of personalized topical antibacterial agent based on fusidic acid already from day 14 significantly reduced the levels of S. aureus and E. coli (p < 0.001 compared to pre-treatment values), which could also be reflected in the reduced number of disease relapses. In the control group, the tendency to decrease the number of these opportunistic bacteria was lower (p < 0.001 compared to the results of the main group). The lower value of the EASI index in the patients of the main group (p < 0.001) is also confirmed.
At the 6th month of therapy, the skin microbiome was restored, as evidenced by a greater share of functional microorganisms (Bifidobacterium, Lactobacillus, and S. epidermidis) in patients of both groups. This may indicate the effectiveness of emollient with probiotic component in the treatment of microbial eczema.
Thus, the personalized selection of topical antibacterial agents can be concluded to contribute to better restoration of the skin layer: the number of relapses in the main group of patients at the 6th month of therapy is 2.5 times less (2 relapses vs. 5). However, in order to reduce the relapse rate to zero, a more comprehensive approach is needed, in particular, personalized use of systemic antibacterial agents.
CONCLUSION
The state of skin microbiome in patients with chronic microbial eczema was assessed considering that the biocenosis of the affected areas was characterized by a high degree of contamination with representatives of opportunistic microflora, while the number of representatives of functional flora was limited. It was found that microflora imbalance contributes to the chronicization of the inflammatory process while increasing the risk of relapses. The standard treatment in combination with personalized selection of topical antibacterial agents resulted in a decrease in the share of disease relapses and normalization of skin microflora composition in the patients of the main group. The obtained data confirm the effectiveness of an individualized approach to treatment, taking into account the peculiarities of the microbial composition of the skin. The results are scientifically significant as they demonstrate the possible optimization of treatment of patients with chronic microbial eczema using WGS, thus revealing prospects for the development of more effective therapeutic strategies. In future studies, the relationship between the skin and gut microbiome (the skin–gut axis) is feasible to investigate for long-term treatment outcome, as their condition and characteristics may play a key role in the pathogenesis of the disease.
1 Russian Society of Dermatovenerologists and Cosmetologists. Clinical Guidelines. Eczema. 2024. 56 p.
2 Russian Society of Dermatovenerologists and Cosmetologists. Clinical Guidelines. Eczema. 2024. 56 p.
3 Ibid.
References
1. Silina LV, Schwartz NE. Skin microbiome in case of microbial eczema (in Russian only). Russian Journal of Clinical Dermatology and Venereology. 2019;18(1):49–55 (In Russ.). https://doi.org/10.17116/klinderma20191801149
2. Hülpüsch C, Rohayem R, Reiger M, Traidl-Hoffmann C. Exploring the skin microbiome in atopic dermatitis pathogenesis and disease modification. J Allergy Clin Immunol. 2024;154(1):31–41. https://doi.org/10.1016/j.jaci.2024.04.029
3. Salem I, Ramser A, Isham N, Ghannoum MA. The Gut Microbiome as a Major Regulator of the Gut-Skin Axis. Front Microbiol. 2018;9:1459. https://doi.org/10.3389/fmicb.2018.01459
4. Widhiati S, Purnomosari D, Wibawa T, Soebono H. The role of gut microbiome in inflammatory skin disorders: A systematic review. Dermatol Reports. 2021;14(1):9188. https://doi.org/10.4081/dr.2022.9188
5. Ahmad F, Alam MA, Ansari AW, Jochebeth A, Leo R, Al-Abdulla MN, Al-Khawaga S, AlHammadi A, Al-Malki A, Al Naama K, Ahmad A, Buddenkotte J, Steinhoff M. Emerging Role of the IL-36/IL-36R Axis in Multiple Inflammatory Skin Diseases. J Invest Dermatol. 2024;144(2):206–224. https://doi.org/10.1016/j.jid.2023.11.004
6. Rozhivanova TA, Polesko IV, Shcherbakova MYu. Modern concepts of the microbiocenosis of the skin and intestine in patients with eczema and metabolic syndrome. Russian Journal of Clinical Dermatology and Venereology. 2015;14(2):11–16 (In Russ.). https://doi.org/10.17116/klinderma201514211-16
7. Kobayashi T, Nagao K. Host-microbial dialogues in atopic dermatitis. Int Immunol. 2019;31(7):449–456. https://doi.org/10.1093/intimm/dxz026
8. Kovaleva YuS, Komkina NG. Microbic eczema — trigger points of influence. Meditsinskiy Sovet. 2023;17(2):37–44 (In Russ.). https://doi.org/10.21518/ms2022-031.
9. Baviera G, Leoni MC, Capra L, Cipriani F, Longo G, Maiello N, Ricci G, Galli E. Microbiota in healthy skin and in atopic eczema. Biomed Res Int. 2014;2014:436921. https://doi.org/10.1155/2014/436921
10. Murzina E, Kaliuzhna L, Bardova K, Yurchyk Y, Barynova M. Human Skin Microbiota in Various Phases of Atopic Dermatitis. Acta Dermatovenerol Croat. 2019;27(4):245–249.
11. Kuznetsov KO, Tukbaeva LR, Kazakova VV, Mirzoeva KR, Bogomolova EA, Salakhutdinova AI, Ponomareva DYu, Garipova AR, Mutsolgova MSM, Galimkhanov AG, Sakhibgareev MI, Guzhvieva ER. The Role of COVID-19 in Antibiotic Resistance in Pediatric Population. Pediatric pharmacology. 2022;19(6):503–513 (In Russ). http://dx.doi.org/10.15690/pf.v19i6.2465
12. Shah RA, Hsu JI, Patel RR, Mui UN, Tyring SK. Antibiotic resistance in dermatology: The scope of the problem and strategies to address it. J Am Acad Dermatol. 2022;86(6):1337–1345. https://doi.org/10.1016/j.jaad.2021.09.024
13. Miller AC, Adjei S, Temiz LA, Tyring SK. Antibiotic Resistance in Dermatology Part 1: Mechanisms of Resistance. Skin Therapy Lett. 2023;28(1):7–10
14. Stracy M, Snitser O, Yelin I, Amer Y, Parizade M, Katz R, Rimler G, Wolf T, Herzel E, Koren G, Kuint J, Foxman B, Chodick G, Shalev V, Kishony R. Minimizing treatment-induced emergence of antibiotic resistance in bacterial infections. Science. 2022;375(6583):889–894. https://doi.org/10.1126/science.abg9868
15. Bin L, Malley C, Taylor P, Preethi Boorgula M, Chavan S, Daya M, Mathias M, Shankar G, Rafaels N, Vergara C, Potee J, Campbell M, Hanifin JM, Simpson E, Schneider LC, Gallo RL, Hata T, Paller AS, De Benedetto A, Beck LA, Ong PY, Guttman-Yassky E, Richers B, Baraghoshi D, Ruczinski I, Barnes KC, Leung DYM, Mathias RA. Whole genome sequencing identifies novel genetic mutations in patients with eczema herpeticum. Allergy. 2021 Aug;76(8):2510–2523. https://doi.org/10.1111/all.14762
16. Olisova OYu, Svitich OA, Poddubikov AV, Vartanova NA, Potapova MB. Microbiological assessment of the effectiveness of standard therapy in atopic dermatitis. Vestnik Dermatologii i Venerologii. 2023;99(3):44–52 (In Russ). https://doi.org/10.25208/vdv1364
17. Fölster-Holst R. The role of the skin microbiome in atopic dermatitis - correlations and consequences. J Dtsch Dermatol Ges. 2022;20(5):571– 577. https://doi.org/10.1111/ddg.14709
18. Tlish MM, Kuznetsova TG, Naatyzh ZhYu, Psavok FA. Microbial eczema: possibilities of correction at the present stage. Vestnik Dermatologii i Venerologii. 2018;94(4):60–67 (In Russ). https://doi.org/10.25208/0042-4609-2018-94-4-60-67
19. Hanifin JM, Baghoomian W, Grinich E, Leshem YA, Jacobson M, Simpson EL. The Eczema Area and Severity Index-A Practical Guide. Dermatitis. 2022;33(3):187–192. https://doi.org/10.1097/DER.0000000000000895
20. Muravyova AS, Lykov IN. Medical and environmental aspects of antibiotic resistance of the skin microbiome in family members. Problemy Regional’noi Ekologii. 2023;3:27–31 (In Russ). https://doi.org/10.24412/1728-323X-2023-3-27-32
21. Piruzyan AL, Niewozinska ZA, Korsunskaya IM. Skin bacterial infections — always a topical problem. Medical Council. 2021;8:63–66 (In Russ.). https://doi.org/10.21518/2079-701X-2021-8-63-66
22. Tarasko AD. Chronic deep recurrent pyoderma in the outpatient practice of the surgeon. Ambulatory Surgery (Russia). 2021;18(2):144–150 (In Russ.). https://doi.org/10.21518/1995-1477-2021-18-2-144-150
23. Tlish MM, Popandopulo EK. Evaluation of the effectiveness of ultratonotherapy in patients with microbial eczema. Problems of Balneology, Physiotherapy and Exercise Therapy. 2022;99(5):48–53 (In Russ.). https://doi.org/10.17116/kurort20229905148
24. Mlynarczyk-Bonikowska B, Kowalewski C, Krolak-Ulinska A, Marusza W. Molecular Mechanisms of Drug Resistance in Staphylococcus aureus. Int J Mol Sci. 2022;23(15):8088. https://doi.org/10.3390/ijms23158088
25. Xie J, Li M, Yang S, Dong Q. Topical administration of mupirocin ointment and fusidic acid in bacterial infection-induced skin diseases. Postepy Dermatol Alergol. 2025;42(1):42–46. https://doi.org/10.5114/ada.2024.145185
26. Chu DK, Chu AWL, Rayner DG, Guyatt GH, Yepes-Nuñez JJ, Gomez-Escobar L, Pérez-Herrera LC, Díaz Martinez JP, Brignardello-Petersen R, Sadeghirad B, Wong MM, Ceccacci R, Zhao IX, Basmaji J, MacDonald M, Chu X, Islam N, Gao Y, Izcovich A, Asiniwasis RN, Boguniewicz M, De Benedetto A, Capozza K, Chen L, Ellison K, Frazier WT, Greenhawt M, Huynh J, LeBovidge J, Lio PA, Martin SA, O’Brien M, Ong PY, Silverberg JI, Spergel JM, Smith Begolka W, Wang J, Wheeler KE, Gardner DD, Schneider L. Topical treatments for atopic dermatitis (eczema): Systematic review and network meta-analysis of randomized trials. J Allergy Clin Immunol. 2023;152(6):1493–1519. https://doi.org/10.1016/j.jaci.2023.08.030
27. Kotsur YuM, Chernykh TF, Flisyuk EV, Narkevich IA. Development of a topical formulation of a thiadiazole derivative. Drug development & registration. 2024;13(4):121–128 (In Russ.). https://doi.org/10.33380/2305-2066-2024-13-4-1931
28. Lax SJ, Van Vogt E, Candy B, Steele L, Reynolds C, Stuart B, Parker R, Axon E, Roberts A, Doyle M, Chu DK, Futamura M, Santer M, Williams HC, Cro S, Drucker AM, Boyle RJ. Topical Anti-Inflammatory Treatments for Eczema: A Cochrane Systematic Review and Network Meta-Analysis. Clin Exp Allergy. 2024;54(12):960–972. https://doi.org/10.1111/cea.14556
About the Authors
V. V. LazarevРоссия
Venyamyn V. Lazarev - Assistant of the Department of Dermatovenerology,
Mitrofana Sedina str., 4, Krasnodar, 350063
M. M. Tlish
Россия
Marina M. Tlish - Dr. Sci. (Med.), Professor (academic title), Head of the Department of Dermatovenerology,
Mitrofana Sedina str., 4, Krasnodar, 350063
M. E. Shavilova
Россия
Шавилова Марина Евгеньевна - кандидат медицинских наук, ассистент кафедры дерматовенерологии,
Mitrofana Sedina str., 4, Krasnodar, 350063
Supplementary files
Review
For citations:
Lazarev V.V., Tlish M.M., Shavilova M.E. Personalized selection of topical antibacterial agents in patients with microbial eczema based on whole genome sequencing data: A prospective сomparative randomized study. Kuban Scientific Medical Bulletin. 2025;32(4):18-32. https://doi.org/10.25207/1608-6228-2025-32-4-18-32
JATS XML








































